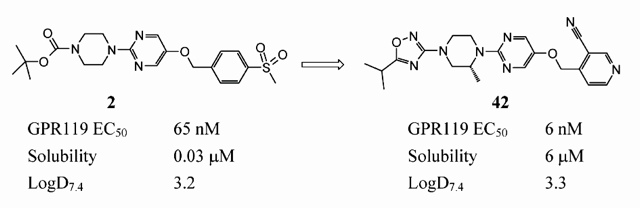
A recent publication from AstraZeneca provides a really interesting overview of the approach that they took to optimise the solubility of a series of GPR119 agonists. The initial lead compound 2 was only poorly soluble but, by obtaining small molecule x-ray structures of this and subsequent compounds, the AZ team were able to identify H-bonding networks within the crystal lattice and to target molecular features with the potential to disrupt these and other intermolecular interactions, thus lowering the melting point and increasing aqueous solubility. The paper describes this process in some detail and also highlights other key strategies that were used to progress from compound 2 through to 42, a clinical candidate for treating type II diabetes. The paper also serves to illustrate and build upon the concepts described in the review that we blogged previously.
-
Join 92 other subscribers
-
Recent Posts
Top Rated
Most Popular:
- Improving Solubility using X-ray crystal structure data
- MDM2-p53 protein-protein interaction inhibitors
- GPCR structure and modeling
- Drug discovery approaches to minimise drug induced liver injury in man
- More physicochemical musings & ChEMBL
- Review: Optimizing CNS access
- Review: Bioisosteres in Drug Design
- Discovery of PF-04691502 (PI3K/mTOR dual inhibitor)
- Synthesis: Sulfonylated Pyridines
- Discovery of MK-1220 (Hepatitis C NS3/4A protease inhibitor)
Blogroll
Conferences
Intellectual Property
Journals
MedChem Resources
News
Organic Chemistry Resources
- Abbott
- adverse events
- allosteric
- allosteric inhibitor
- amine basicity
- Analgesic
- animal PK
- anti-viral
- AstraZeneca
- Aurora
- beta-secretase
- bioactivation
- BIRB-796
- Boehringer Ingelheim
- cancer
- cardiotoxicity
- cellular potency
- CGRP
- chloro
- CNS penetration
- computational method
- crystal packing
- CYP450
- Deuterium
- Doramapimod
- FAAH
- Fatty acid amide hydrolase
- fluorine
- free drug
- functional antagonist
- Gly-T1
- gpcr
- hepatotoxicity
- hERG
- immunosuppressant
- isostere
- kinase
- kinetic isotope effect
- Lexicon Pharmaceuticals
- LipE
- Merck
- metabolism
- mTOR
- nitrile
- Novartis
- osteoarthritis
- P-gp
- p38
- Pain
- PF-04457845
- PF-3845
- Pfizer
- Phase I
- Phase II
- Phase III
- POC
- PPB
- protein binding
- PSA
- reactive metabolite
- review
- RG1678
- rheumatoid arthritis
- Roche
- S1P1
- S1PL
- Schizophrenia
- solubility
- Structural alert
- Structure Based Design
- switch pocket
- synthesis
- toxicity
- Transporters
- X-ray data
Categories


MCB! Yes!
🙂 nice to know someone missed me!
CJ isn’t the only one! Nice to see a new and interesting post, keep it up…
nice
Reblogged this on medchemhotsun.
Will the obtaining of molecule x-ray structures will influence the performance of the compound?-BOC Sciences